Dr Harald Putzer – Rna Metabolism: How Different Bacteria Reached The Same Conclusion In Their Own Ways
Dr Harald Putzer at the Institut de Biologie Physico-Chimique, France, investigates how bacteria regulate the expression of genes by degrading their transcripts with RNase enzymes, and how organisms separated by billions of years of evolution, figured out how to do it with remarkably similar techniques.
It is reasonable to call it common knowledge that the blueprints for building a person are stored within our cells in the form of DNA. With slight variations, the basic mechanisms of how DNA is ‘read’ to produce the proteins which it codes for, are conserved in all of life’s domains, from bacteria to complex multicellular organisms like animals and plants. One of DNA’s most useful properties is its stability. DNA could be considered the master copy of the instructions, which are not read directly by the protein-building machinery (ribosomes). Instead, DNA is transcribed into a similar molecule called RNA, which is much less stable. RNA which is used in this way is called ‘messenger RNA’ or mRNA, to distinguish it from other types of RNA within the cell. If DNA is the master copies of the blueprints, then mRNA could be considered the photocopies taken to the building site.
It is the mRNA which is read by ribosomes, as instructions for putting together chains of specific amino-acids – the building blocks of proteins. Using mRNA in this way allows more finely tuned regulation of the amount of protein ultimately produced from a given gene. Each gene often exists as a single copy within the genome, and if a large amount of protein is required, which can change from moment to moment depending on the needs of the cell, then multiple copies of the gene can be produced in the form of mRNA. Each copy allows the production of multiple protein molecules as it is read by the ribosomes.
When the cell decides to stop producing a particular protein, and suspend further production of mRNA, the relevant mRNA molecules already present can be rapidly degraded and their constituent parts recycled. In bacteria this degradation can take place in a matter of seconds or in over an hour, depending on the specific mRNA molecule. ‘In contrast to DNA which is a very stable, mRNA is a very fragile molecule that only survives a few minutes in a bacterial cell,’ Dr Putzer explains. Bacteria are thought to regulate this process most efficiently by using an ‘all-or-none’ pattern, where control is implemented at the initiating step within the degradation process. Degradation of mRNA is carried out by a particular type of enzyme, called RNase. The many types of RNase vary in their specific functions, such as in their mRNA targets or whether they degrade mRNA from the terminal ends of the molecule (an exonuclease) or cleave the molecule somewhere between these termini (an endonuclease).
The interests of Dr Putzer and his group are concerned with the structure, function and evolution of this class of proteins and how they work to regulate gene expression and RNA metabolism.
Evolutionary distance
As humans, we might consider ourselves highly distinct from things such as plants, yeast, or single-celled amoebae, but we have a common biological feature which is absent in bacteria – a cell nucleus. The nucleus evolved around 2 billion years ago, but long before this (about 3 billion years ago) the evolutionary paths of the two major types of bacteria, gram negative and gram positive, diverged from each other. One of the defining differences between these bacteria is the structure of their cell walls, but the point is that in terms of evolutionary timescales, they are more distinct from each other than a human is from a mushroom. Two of the most highly studied representatives of these classes of bacteria are Escherichia coli, a gram negative and probably the most heavily studied organism in biology, and Bacillus subtilis, a gram positive. ‘I started to work with B. subtilis because it became clear quite quickly that even among bacteria regulation of gene expression can involve very divergent mechanisms,’ Dr Putzer tells us. ‘The large evolutionary distance between E. coli and B. subtilis has been very rewarding in terms of exploring new mechanisms and enzymes involved in gene control.’
Both organisms contain RNase enzymes, but they are evolutionarily so distinct from each other that they could scarcely be said to be comparable at the DNA sequence level. In recent years, the work of Dr Putzer and his lab has shown that RNase E (from E.coli) and RNases Y and J, found in B. subtilis, have evolved – independently – highly similar structural properties and mechanistic features.
‘We focus on the structural and functional study of RNase Y, a key ribonuclease in B. subtilis. Together with RNase J and RNase E these three enzymes probably represent the major ribonucleases that determine the strategies used by bacteria to initiate mRNA decay.’
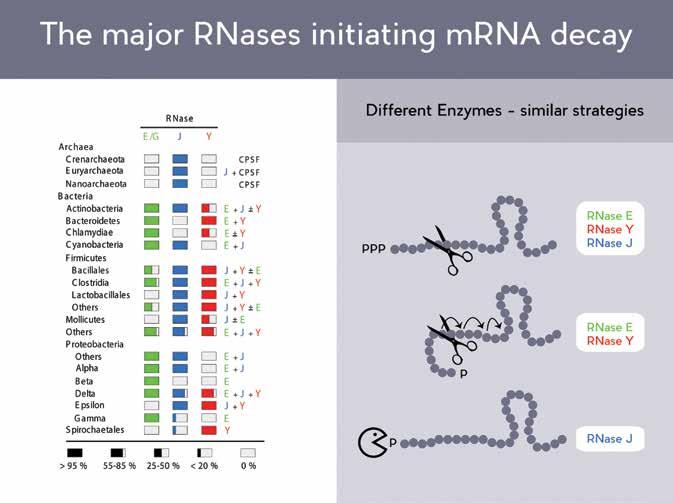
Different enzymes – similar strategies
In 2008, the Putzer lab elucidated the mechanism of function of the first RNase known to possess both endonuclease and exonuclease activity. The team has also demonstrated that these enzymatic activities are carried out by the same catalytic site within the protein. Though versions of this protein are present in throughout the bacterial kingdom, it is notably absent from E. coli. Structurally, RNase J and RNase E appear quite similar, despite a complete absence of similarity in the actual DNA sequences of the two genes which code for them.
A year later, the group published research into the role of another important RNase, called RNase Y. This enzyme is found in B. subtilis, as well as around 40% of other bacteria, and shows a similar effect to RNase E, on the total amount of mRNA within the cell. Rather than only degrading a small number of specific mRNA targets, these enzymes have wide ranging mRNA targets, having a major reductive effect on the total quantity of mRNA. Another similarity between these two enzymes lies in their propensity to interact with multiple, distinct enzymes, to form a complex of RNA and protein degrading enzymes, called a degradosome.
The discovery of RNase Y led to a reevaluation of RNA decay models in B. subtilis, which until then had focussed predominantly on the exonuclease activity of RNase J, while RNase Y strongly affects the total mRNA of the cell through its endonuclease activity. The endonuclease specificity of RNases J and Y are remarkably similar to that of RNase E, with all three showing a strong preference for targeting mRNA molecules which have been processed in a particular way, by the addition of a phosphate molecule at one end of the mRNA molecule.
The influence of a particular enzyme on a cell can also be modulated by its precise location inside the cell. Bacterial cells are not compartmentalised by the presence of internal membranes, unlike in eukaryotes. Regardless of this, some internal cellular components are still localised with some precision. RNase Y appears to show a similar pattern of localisation within the cell, as that seen for RNase E in E. coli. Both enzymes are found at the periphery of the cell, tethered to the cell membrane, though the reason for this pattern of localisation is not clear.
These shared properties make it likely that the way mRNA is processed and degraded are more similar between these two evolutionarily diverged classes of organisms – gram positive and negative – than previously thought.
 The effects of RNase Y
The effects of RNase Y
As mentioned before, mRNA is only one of the types of RNA molecules present in the cell. RNA has numerous functions, often also involving regulation of gene expression, when it takes the form of ‘non-coding RNA’ – RNA that does not directly code for a protein. RNase Y is also thought to have a role in processing these non-coding RNA molecules. In experiments involving strains of bacteria where the amount of RNase Y is depleted, the specific mRNA and non-coding RNA molecules can be measured and compared to those found in a strain with normal levels of functional RNase Y. In these experiments, around 1600 mRNA transcripts are found in the RNase Y depleted strain, which are upregulated in comparison to when the enzyme in present, while several hundred non-coding RNA molecules are also found to be increased in its absence. Interestingly, across several studies, though the numbers of these molecules seem to be in agreement, the actual identity of the transcripts is highly variable.
The variability in the activity of RNase Y seems to be at least partly dependent on growth phase of the bacteria. Microorganisms show highly different patterns of gene expression based on whether they are rapidly dividing (exponential phase), such as they would do in ideal environmental and nutritional conditions, or if they are in a stationary phase, where their overall numbers are steady. Studies looking at the activity of RNase Y in pathogenic gram positives, such as Staphylococcus aureus (the ‘SA’ in MRSA) have shown that during exponential growth, the number of genes affected by depletion of RNase Y is only around 100 – far less than in Bacillus subtilis – but that this number is greatly increased during stationary phase.
Another experiment in Staphylococcus aureus artificially altered the localisation of the enzyme, from being on the membrane to floating around freely inside the cell, but found that this did not have much of an effect on the activity or specificity of the enzyme, at least during exponential growth. In B. subtilis however, Dr Putzer’s group have found preliminary data suggesting that at least for certain transcripts, the enzyme’s localisation at the cell membrane is essential for it to degrade them effectively.
The future of RNase research
Dr Putzer and his team plan to continue his laboratory’s research into RNase Y and RNase E, by using a wide variety of approaches. Though the structure of RNase J is known to be similar to that of RNase E, the structure of RNase Y is still unknown, and the group is currently working towards determining it. He plans to sequence RNA across the whole genomes of these bacteria, in order to determine which ones are targeted specifically by these RNases. Another future aim of his laboratory is to study how interacting proteins of RNase Y, such as those involved in biofilm formation, modify the activity of the enzyme. His lab will also take advantage of recent advancements made in the field of super-resolution microscopy, which offers resolution beyond the theoretical limit of a light microscope, therefore allowing single RNA molecules to be imaged within the cell. Using superresolution microscopy in live cells might help to answer questions about the patterns made by RNase E and RNase Y as they move along the inner cell membrane, as well as how they interact with other components of the degradosome as it assembles and disassembles. Dr Putzer hopes to use these techniques to learn more about the commonalities and differences between the mRNA degradation strategies in these two evolutionarily distinct organisms. ‘Answers to these questions might help shed light on some fundamental aspects of mRNA metabolism that might be important to all bacteria,’ he explains.
This research is fundamentally important to understand how essential biological processes are conserved between highly divergent bacterial species, and will have impacts on agricultural, industrial and biomedical research.
Meet the researcher

Dr Harald Putzer
Head, Microbial Gene Expression Department
CNRS UMR8261 – Institut de Biologie Physico-Chimique
Université Paris Diderot, France
Dr Harald Putzer graduated from the University of Innsbruck, Austria in 1980, with a Master’s degree in Natural Sciences. He then went on to complete a PhD at the Max Planck Institute of Biochemistry, in Germany, studying how light can regulate gene expression in slime molds. Dr Putzer then moved to Paris, where he performed research at the Institut de Biologie Physico-Chimique, largely focusing on the study of gene regulation in bacteria. Here, he currently holds the position of Head of the Microbial Gene Expression Department.
CONTACT
T: (+33) 1 58 41 51 27
W: http://www.ibpc.fr/UMR8261/equipePutzer/HomePutzer.htm
KEY COLLABORATORS
Dr A. Carpousis, University Paul Sabatier, Toulouse, France
Dr FA Wollmann, Institut de Biologie Physico-Chimique, Paris, France
FUNDING
CNRS, Labex Dynamo and Agence Nationale de la Recherche (ANR)
REFERENCES
S Laalami, L Zig and H Putzer, Initiation of mRNA decay in bacteria, Cell Mol. Life Sci., 2014, 71, 1799–1828.
K Shahbabian, A Jamalli, L Zig and H Putzer, RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis, The EMBO Journal, 2009, 28, 3523–3533.
IL de la Sierra-Gallay, L Zig, A Jamalli and H Putzer, Structural insights into the dual activity of RNase J, Nature Structural and Molecular Biology, 2008, 15, 206–212.


