Dr Bert Lampson – Bacteria: Deadly but Useful
Bacteria are found everywhere – in humans, animals and the environment – and are best known for being able to cause painful and even fatal infections. It may come as surprise, therefore, to learn that bacteria can also have useful applications. The lifetime work of Dr Bert Lampson at East Tennessee State University in the USA has focussed on the dangers and applications of bacteria. Over his career, Dr Lampson has discovered new mechanisms of antibiotic resistance, novel antibiotics, and bacterial proteins, with important applications for science and healthcare.
Revealing Antibiotic Resistance Mechanisms
Dr Bert Lampson began his research career as a graduate student in the early 1980s while studying at the University of Missouri-Columbia School of Medicine. At this time, scientists were developing new research technologies, including DNA sequencing. This process involves determining the sequence of nucleotides, the building blocks of DNA. The sequences can tell us what genes bacteria carry. Genes are inherited by bacteria and have many functions, such as encoding proteins that are used within the bacterial cell.
Dr Lampson used DNA sequencing to discover antibiotic resistance genes that are carried by plasmids. Antibiotic resistance genes are genes that encode mechanisms that lead to bacteria becoming resistant to antibiotics. Furthermore, plasmids are small circles of DNA that are physically separate from chromosomal DNA. The chromosomal DNA contains a large number of genes required to maintain cellular functions. In contrast, plasmids can carry a small number of additional genes, also known as accessory genes, that can help bacteria to adapt and survive in their environments. Furthermore, plasmids can be transferred amongst bacteria populations and spread genes. This has led to the spread of antimicrobial resistance genes that prevent antibiotic drugs from successfully treating an infection.
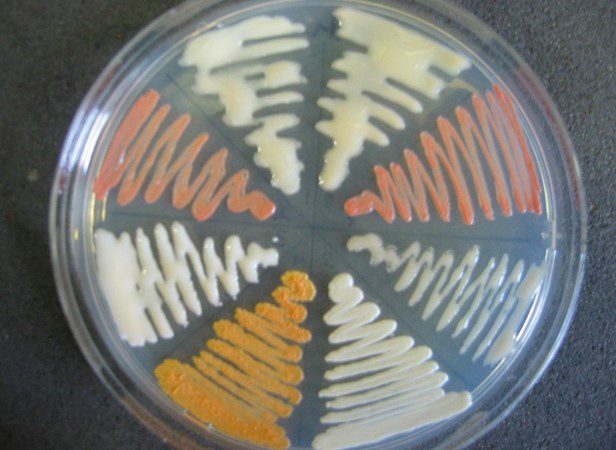
‘Wheel’ plate containing different Rhocococcus bacteria. Credit Bert Lampson.
The early work of Dr Lampson first involved sequencing a previously discovered plasmid in Staphylococcus epidermis. This bacterium is found around naturally on the skin. However, when the skin is broken the bacteria can enter the body and cause harmful infections. This particularly occurs when medical equipment, such as needles, are inserted into the body. These infections are most often treated with antibiotics.
Dr Lampson, under the guidance of Dr Joseph Parisi, showed that plasmids can induce resistance to three antibiotics (macrolides, lincosamide and streptogramin). This was demonstrated using transduction (gene transfer between bacteria using a bacterial specific virus called a phage), whereby the plasmids carrying the resistance genes were input into the bacteria not carrying the plasmid. Dr Lampson and his team then treated two sets of bacteria, those with and those without the plasmid, with antibiotics to see if carrying the plasmid led to bacteria becoming resistant.
Using DNA sequencing Dr Lampson and his fellow researchers pinpointed resistance to be due to a new gene, ermM. This gene has a similar sequence to a previously described resistance gene. However, the team observed changes upstream of the gene sequence, leading to changes in how the gene is expressed. ermM is constantly ‘switched on’, meaning it is continually expressed. The gene encodes an enzyme, a protein that is able to help the cell to carry out reactions. Critically, the protein alters the binding site, that is, the target site for antibiotics and prevents them from working.
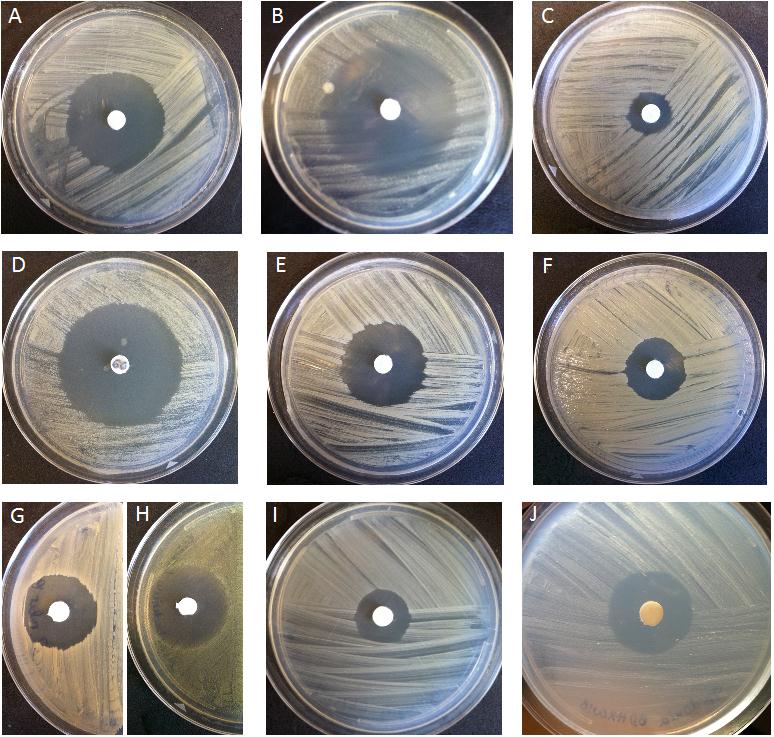
Spectrum of activity of the novel antibiotic from the Rhodococcus strain. Credit R. Borisova.
Remarkably, Dr Lampson also discovered a novel resistance mechanism that is carried by plasmids in the same species of bacteria. The newly discovered mechanism led to resistance to erythromycin. This is an antibiotic that is used to treat a number of infections. It operates by preventing bacterial cells from producing proteins that are essential for life, as well as replication.
Dr Lampson and his colleagues identified a new resistance gene that they designated erpA that was carried on a plasmid pNE24. Experiments that can detect specific DNA sequences, known as southern blot hybridisation, showed that the plasmid carries a new resistance mechanism. This is because the plasmid contains different DNA sequences to other plasmids that also carry erythromycin resistance genes. The new gene makes the bacterial cell less permeable, preventing the uptake of the antibiotic. This was shown by labelling the antibiotic with a radioactive carbon (Carbon-14) which helps scientists to detect the amount of antibiotic that the bacterial cells uptake.
This research holds important implications for understanding how bacteria become resistant to antibiotics and how this can spread amongst bacterial populations, particularly in hospitals.

Searching for Novel Antibiotics
When Dr Lampson started his role as a Research Scientist at East Tennessee State University (ETSU), he wanted to work on antibiotics again. However, this time, he took inspiration from work he had completed with the energy company, Energy Biosystems. This company was using an interesting soil bacterium, Rhodococcus, to try and produce cleaner fuels but Dr Lampson saw an additional opportunity for this research. Although ETSU is not a primarily research university, Dr Lampson was also keen for students to participate in this exciting new research project.
This work focused on exploiting Rhodococcus for antibiotics, which at first, may seem paradoxical. Why would bacteria produce antibiotics that are used to kill them? The reason is that species of soil bacteria have to compete with one another to survive. Therefore, bacteria produce antibiotics to outcompete other species.
Dr Lampson and his students grew Rhodococcus on plates that contain agar, on which bacteria can survive. They extracted molecules from these agar plates and then they searched for molecules and proteins from Rhodococcus using different forms of chromatography. This process helps to separate the different molecules from one another. After separating the extracts, the team coated small disks with the extracts and tested whether the bacteria could survive in its presence.
This work led to the discovery of a new compound that could inhibit the growth of bacteria with potentially significant applications in medical and veterinary practice. As such, this key study resulted in several more research projects, and towards the end of 2018, a publication for Dr Lampson and his students in the high impact journal PLoS ONE.

Discovering the First Bacterial Reverse Transcriptase
Dr Lampson has additional research interests outside the field of antibiotic resistance. Notably, whilst working as a postdoctoral researcher at the Robert Wood Johnson Medical School, he discovered a protein with very important applications.
Using soil bacteria, this time Myxococcus, Dr Lampson discovered the first example of a bacterial reverse transcriptase. This is a protein, also known as an enzyme, that is responsible for forming DNA from RNA. Generally, people associate DNA with producing RNA and RNA with making proteins. However, this is a simplified view – in reality, the process is much more complex.
Reverse transcription is the formation of DNA from RNA and this takes place in most organisms, including humans and bacteria. Interestingly, it is believed that reverse transcription was a key process in the origins of life and it is hypothesised that RNA was the source of genetic information and cellular functions. This collectively is known as the ‘RNA world’.
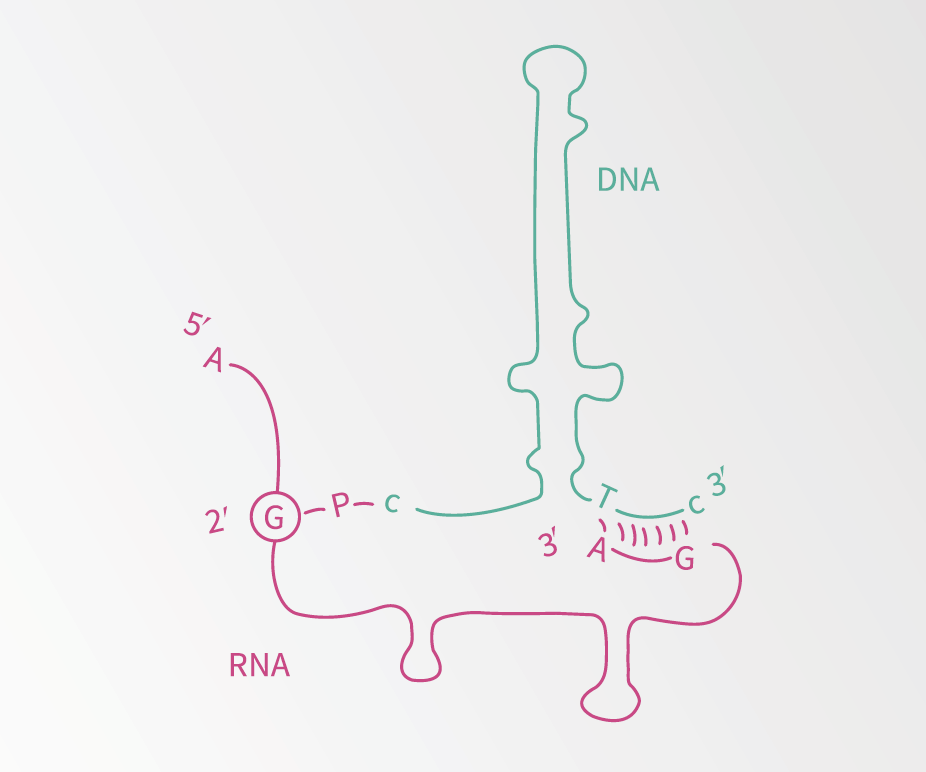
Illustration of msDNA.
Dr Lampson, under the mentorship of Professor Masayori Inouye, discovered bacterial reverse transcriptase after the discovery of another strange bacterial element, msDNA. This molecule contains both RNA and DNA bonded together. Dr Lampson and his colleagues showed that this molecule was produced by reverse transcriptase, making this the first reverse transcriptase to be identified in bacteria. Importantly, reverse transcriptase can be used to make DNA from RNA, which in turn means the DNA can be identified using a process known as polymerase chain reaction (PCR) or DNA sequencing. This allows scientists to figure out what DNA is being expressed.
These findings further led to the discovery of a thermostable reverse transcriptase that can operate at high temperatures. High temperatures otherwise alter the structure of a protein and prevent it from functioning. Although reverse transcriptase had been identified from viruses previously, heat stable bacterial reverse transcriptases are more advantageous because they increase the accuracy of DNA synthesis and are potentially more stable under laboratory experimental conditions.
Dr Lampson and his team searched for the genome sequence of Bacillus stearothermophilus. This bacterium is thermophilic, meaning it can survive at extreme temperatures. They encountered a gene that is similar to genes encoding group II introns that have reverse transcriptase activity, and also provided supportive evidence for the ‘RNA world’ hypothesis. After the scientists cloned the gene and input into Escherichia coli to produce the protein, they confirmed it has reverse transcriptase activity and could operate at temperatures as high as 75oC. Dr Lampson and ETSU have now patented the reverse transcriptase and have licensed it to a company called InGex that developed it as a commercially available enzyme called TGIRT III.

Dr Lampson continues his research and lecturing at ETSU. His current research largely focusses upon looking at the metabolism of soil bacterium Rhodococcus and potential antibiotics produced by this bacterium. He also continues to take interest in reverse transcriptase and their diversity amongst bacteria. Collectively, his work shows that although bacteria can cause harmful, resistant infections, they also harbour proteins with useful applications that can be exploited and bring important benefits.
Reference
https://doi.org/10.33548/SCIENTIA630
Meet the researcher

Dr Bert Lampson
Department of Health Sciences
East Tennessee State University
Johnson City
Tennessee, TN
USA
Dr Bert Lampson received his PhD in Medical Microbiology from the University of Missouri-Columbia School of Medicine in 1986. He then undertook postdoctoral research in the laboratory of Dr Masayori Inouye at the University of Medicine and Dentistry, Robert Wood Johnson Medical School. In 1998, Dr Lampson became an Assistant Professor in the Department of Health Sciences at East Tennessee State University which was followed by promotion to his current role of Associate Professor in 2004. He has had a successful research career spanning four decades, during which time he has contributed significantly to tackling the worldwide problem that is antibiotic resistance.
CONTACT
E: lampson@etsu.edu
KEY COLLABORATORS
Dr Masayori Inouye, Robert Wood Johnson Medical School
Dr Abbas Shilabin, East Tennessee State University
Professor Joseph T. Parisi (deceased)
Graduate students: J. Vellore, S. Moretz, T. Barber, M. Carr, R. Borisova, A. Ward and K. Sellick
FUNDING
Takara Bio
East Tennessee State University Research Foundation
Department of Health Sciences, ETSU
FURTHER READING
AL Ward, P Reddyvari, R Borisova, AG Shilabin, BC Lampson, An inhibitory compound produced by a soil isolate of Rhodococcus has strong activity against the veterinary pathogen R. equi, PLoS ONE, 2018, 13(12), e0209275.
BC Lampson, M Inouye, S Inouye, Retrons, msDNA and the bacterial genome, Cytogenetic and Genome Research, 2005, 110, 491–499.
J Vellore, SE Moretz, BC Lampson, A group II intron-type open reading frame from the thermophile Bacillus (Geobacillus) stearothermophilus encodes a heat-stable reverse transcriptase, Applied and Environmental Microbiology, 2004, 70, 7140–7147.
JT Parisi, J Robbins, BC Lampson, HW Hecht, Characterization of a macrolide lincosamide, and streptogramin resistance plasmid in Staphylococcus epidermidis, Journal of Bacteriology, 1989, 148, 559–564.
BC Lampson, JT Parisi, Naturally occurring Staphylococcus epidermidis plasmid expressing constitutive macrolide-lincosamide-streptogramin B resistance contains a deleted attenuator, Journal of Bacteriology, 1986, 166, 479–483.
BC Lampson, JT Parisi, Nucleotide sequence of the constitutive macrolide-lincosamide-streptogramin B resistance plasmid pEN131 from Staphylococcus epidermidis and homologies with Staphylococcus aureus plasmids pE194 and pSN2, Journal of Bacteriology, 1986, 167, 888–892.
BC Lampson, W von David, JT Parisi, A novel mechanism for plasmid-mediated erythromycin resistance is conferred by pNE24 from Staphylococcus epidermidis, Antimicrobial Agents and Chemotherapy, 1986, 30, 653–658.

Want to republish our articles?
We encourage all formats of sharing and republishing of our articles. Whether you want to host on your website, publication or blog, we welcome this. Find out more
Creative Commons Licence
(CC BY 4.0)
This work is licensed under a Creative Commons Attribution 4.0 International License. 
What does this mean?
Share: You can copy and redistribute the material in any medium or format
Adapt: You can change, and build upon the material for any purpose, even commercially.
Credit: You must give appropriate credit, provide a link to the license, and indicate if changes were made.
More articles you may like
Improving Food Safety of Hydroponic Leafy Greens
Hydroponic farming is experiencing rapid growth worldwide, offering a sustainable and efficient method of producing fresh, nutrient-rich crops. However, the unique conditions of hydroponic systems also present complex food safety challenges. Dr Sanja Ilic and Dr Melanie Lewis Ivey, researchers at The Ohio State University, are at the forefront of efforts to understand and mitigate the risks of human pathogen contamination in commercial hydroponic production. Their pioneering work is providing crucial insights and practical guidance to help ensure the safety and nutritional value of hydroponically grown leafy greens.
Dr Paul Robertson | Artificial Intelligence in the Cockpit: New Systems Could Help Prevent Aviation Accidents
Despite significant advances in aviation safety over recent decades, accidents still occur that could potentially be prevented with better warning systems. Dr Paul Robertson of Dynamic Object Language Labs, Inc. (DOLL) is leading groundbreaking research into how artificial intelligence could help pilots avoid dangerous situations. His team’s work reveals promising developments and important cautions about implementing AI in aircraft cockpits, with implications for the future of aviation safety.
Dr Yasjka Meijer | Monitoring Greenhouse Gas Emissions from Space: The Copernicus CO2M Mission
Atmospheric concentrations of carbon dioxide (CO2) and methane (CH4) have been steadily rising due to human activities, contributing to global climate change. Dr Yasjka Meijer from the European Space Agency is responsible for the objectives and requirements of the Copernicus Anthropogenic Carbon Dioxide Monitoring (CO2M) mission – a constellation of satellites that will enable the monitoring of anthropogenic greenhouse gas emissions from space with unprecedented accuracy and detail. This groundbreaking mission aims to support international efforts to reduce emissions and combat climate change.
Dr Silvia Remeseiro | Mapping the Epigenetic Landscape of Glioblastoma Progression
Glioblastoma, the most aggressive form of brain cancer, continues to challenge medical professionals with its poor survival rates. Recent groundbreaking research by Dr Silvia Remeseiro and her colleagues at Umeå University in Sweden has shed light on the complex epigenetic and chromatin-related mechanisms underlying the communication between neurons and glioma cells. This research opens new avenues for understanding and potentially treating this formidable disease.




