Professor Charles Bangham – The Forgotten Cousin of HIV
The catastrophic impact of HIV – human immunodeficiency virus – is well-known worldwide. Not so well known is its distant cousin HTLV-1, human T cell leukaemia virus type 1, which also has the potential to destroy lives. The study of HTLV-1 can shed light on HIV itself. A team at Imperial College London, led by Professor Charles Bangham, has been working to raise awareness of this under-researched virus and to find better treatments for the devastating illnesses that it causes.
A Global Problem That Is on the Move
An estimated 10 million people worldwide are infected with the human T cell leukaemia virus type 1 (HTLV-1). About one in ten of them eventually develop adult T cell leukaemia, a cancer of the blood cells, or another condition called HTLV-associated myelopathy, characterised by progressive lower limb weakness and paralysis. These diseases typically emerge many years after initial infection. Neither disease is curable, both are debilitating, and the leukaemia is usually fatal despite treatment.

Colorized scanning electron micrograph of a T lymphocyte. Credit: NIAID
HTLV-1 infection is most prevalent in sub-Saharan Africa, South America and the Caribbean, Iran, Japan, and Australia. The virus is transmitted through breastfeeding, sexual intercourse, or infusion with contaminated blood products, and is therefore mainly transmitted within families and local communities. However, as travel becomes easier, and people move around more, it is becoming harder to identify those who carry the virus as they may no longer live in a high-risk area. For this reason, it is becoming increasingly difficult to prevent further spread of the virus.
Professor Charles Bangham, Head of the Division of Infectious Diseases at Imperial College London, argues that with HTLV-1 infection becoming more geographically spread there is an urgent need to raise awareness of it among both medical professionals and the public. Professor Bangham and his research team are committed to raising the profile of HTLV-1 and to finding more effective treatments for those coping with the long-term consequences of infection.
HTLV-1 Latency: A Mysterious Balance
A person becomes infected with HTLV-1 when their cells come into contact with the cells of an infected person, for example, an infant during breastfeeding by an HTLV-1-infected woman. When this happens, the virus is able to cross from one cell to the other and enter the cells of the previously uninfected person. HTLV-1 is most likely to infect the T cell, a certain type of white blood cell.
Every cell in the human body holds a complete copy of an individual’s DNA, the genetic code that contains the instructions for making all the proteins needed during life. HTLV-1 is a retrovirus, which means that it incorporates its own DNA into the genetic code of infected cells, causing them to make proteins wanted by the virus. These viral proteins cause the infected cells to appear different from their uninfected peers and thus make them targets for destruction by immune-response cells. HTLV-1 infection also increases the rate in which T cells multiply, so the number of infected cells present at any one time is a balance between the production of new T cells and their destruction by the immune response.
Following infection with HTLV-1, it is usually decades before the symptoms of adult T cell leukaemia or HTLV-associated myelopathy develop, and up to 90% of those infected never become ill at all. One question that continues to challenge researchers is how the virus is able to maintain a population of infected cells in the body for so long, in the face of strong attacks from the immune system.
It was previously thought that the HTLV-1 virus lies dormant in T cells for the decades between infection and disease, during which time its DNA is present in the infected T cells but not used to make proteins. But Professor Bangham’s research team found that the immune cells that attack infected T cells are continuously active during this time, so the infected cells must be making viral proteins in order for them to become a target.
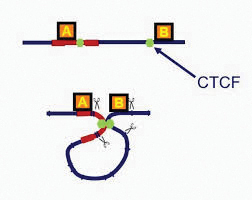
The DNA of the HTLV-1 virus is able to bind to a protein known as CTCF, involved in forming the loops in DNA. Credit Charles Bangham.
What, then, do infected T cells spend their time doing? The team at Imperial College investigated the viral DNA in cells from HTLV-1-infected patients, to measure which genes were being expressed and when. Before a protein can be generated from a piece of DNA, a copy of that section of genetic code is produced, called messenger RNA, and this is then used by the cell’s building machinery to generate the corresponding protein.
To measure which genes infected cells were producing, the research team generated fluorescently labelled probes for the messenger RNA of different HTLV-1 viral genes and used a microscope to visualise the production of this RNA in individual cells. This method provides much more information than those which only look at the average expression across a large number of cells. It has shown that the HTLV-1 gene that codes for a protein called Tax is expressed at very high levels but only intermittently in each infected cell. So only a small proportion of HTLV-1 positive cells, between 1 and 10 per cent, produce Tax at any one time. Critically, Tax marks infected T cells for destruction by the immune system.
This intermittent production of Tax results in an immune response that is continuous, but too weak to significantly reduce the overall number of infected cells. Only a small proportion of the population of cells are attacked at any one time, so the rest are allowed to multiply unhindered. The next step in understanding the virus will be to establish if the same process occurs in HTLV-1-infected cells in patients as in those grown in a dish. Professor Bangham plans to do this by rapidly analysing cells immediately after they have been taken from the patient’s bloodstream.
This finding provides crucial new information on the interaction between HTLV-1 and the immune system. This increases our current understanding by helping identify the parts of the process that could be altered by therapeutics, perhaps, for example, to help the immune system fight infected T cells more effectively.
From HTLV-1 Infection to Leukaemia: Filling in the Gaps
Another unsolved question in the HTLV-1 puzzle is how infection with the virus can lead to cancer and a range of immune diseases. To try to answer it, Professor Bangham and his team have been investigating the effect HTLV-1 has on the genetic code of the infected person.
For a strand of DNA to make a protein it first needs to be unpackaged and made accessible. Viruses such as HTLV-1 and HIV enter cells and insert their own small segment of DNA into the genetic code of the person they have infected. The challenge for medical science is to understand the chain of events, from the insertion of this viral DNA into the infected person’s genetic code onwards, that leads to the later development of symptoms.
The DNA that contains this code – the human genome – is not randomly scattered about the cell but is carefully coiled and folded into loops in a small compartment, called the nucleus. This DNA folding or looping is controlled by the binding of proteins to different sections of the DNA. The way in which this looping takes place is important, as it controls how easily different parts of the genome can be accessed and used by the cell to make new proteins.
Professor Bangham and his team recently found that the DNA of the HTLV-1 virus is able to bind to a protein known as CTCF, which is involved in forming the loops in DNA. This suggested to them that the addition of viral DNA into an infected person’s genetic code might alter the way their DNA is arranged in the nucleus, therefore leading to later disease. To test this, they screened for contact points between the infected genetic code and the incorporated virus DNA. They looked for points that were close in 3D space but according to the sequence of the genetic code should be far apart, thus indicating that the DNA was forming a loop.
The researchers found that the viral DNA was indeed creating loops in the human genome that would not ordinarily be there. They then found that this abnormal DNA looping changes the expression of human genes near the viral DNA, and so can lead to abnormal production of proteins. If this results in the production of a cancer-causing protein, it could explain how HTLV-1 is able to cause adult T cell leukaemia.
The research team showed that, in addition to disrupting the genes in the immediate vicinity of its insertion, the viral DNA is able to disrupt the expression of much more distant stretches of the infected person’s DNA. This is an important finding because it suggests that researchers may need to look further afield to identify the disrupted genes that cause adult T cell leukaemia in HTLV-1 positive patients, which is necessary to inform the ultimate goal of developing effective treatments.
Knowledge Saves Lives
Despite the large number of people infected with HTLV-1, and the likelihood that the number will continue to rise due to our increasingly mobile global population, little effort has been made by governments and health organisations to identify infected people and to limit transmission. There are currently no vaccines or effective therapies, and only limited methods are available for diagnosis and these are usually only used for research purposes.
Scientists working on HTLV-1 see themselves as having wider responsibilities in future beside continuing their research, to raise awareness of the virus both in at-risk populations and in the medical community. These kinds of initiatives have worked in the past. In Japan, for example, pregnant women have been tested for HTLV-1 infection since the 1990s. Women who test positive for HTLV-1 are given information on the virus and counselled to avoid breastfeeding. This has lowered the rate of transmission of HTLV-1 down the family line in this population. However, there remain many at-risk populations in other countries where no such services are offered.
Moving Forward with the HTLV-1 Taskforce
Professor Bangham is a member of the HTLV-1 Taskforce, which was set up in 2014 by the Global Virus Network to push forward the search for new methods of prevention and treatment of HTLV-1 associated diseases, and to promote basic research into the function of the virus. The work of the Taskforce and of Professor Bangham and his team is critical in addressing the societal and scientific challenges presented by HTLV-1.
Meet the researcher

Professor Charles Bangham
Department of Medicine
Imperial College London
London
United Kingdom
Professor Charles Bangham qualified in Medical Sciences at the University of Cambridge and in Clinical Medicine at the University of Oxford, then spent several years working in hospital medicine. In 1987 he completed his PhD on the immune response to respiratory syncytial virus, based at the Medical Research Council National Institute for Medical Research (London) and the University of Oxford. Professor Bangham has continued his study of the immunology and virology of persistent virus infections, particularly HTLV-1, and has published 141 primary research papers in this area. Professor Bangham is now Head of the Division of Infectious Diseases at Imperial College, London. He is a Wellcome Trust Investigator, recipient of multiple Medical Research Council Project Grants, and is a Fellow of the UK Academy of Medical Sciences. He was awarded the BMC Retrovirology Prize in 2007 and the International Retrovirology Association Basic Science Prize in 2015.
CONTACT
W: https://www.imperial.ac.uk/people/c.bangham
KEY COLLABORATORS
Professor Ewan Birney, European Bioinformatics Institute, Cambridge, UK
Professor Masao Matsuoka, Kumamoto University, Japan
Professor Yorifumi Satou, Kumamoto University, Japan
Professor Chris Schofield, Oxford University, UK
Professor Graham Taylor, Imperial College London, UK
FUNDING
Wellcome Trust Senior Investigator Award (2013–2018)
Medical Research Council Project Grant (2013–2018)
Wellcome Trust Investigator Award (2018–2022)
Medical Research Council Project Grant (2018–2020)
FURTHER READING
A Melamed, H Yaguchi, M Miura, A Witkover, TW Fitzgerald, E Birney, CRM Bangham, The human leukemia virus HTLV-1 alters the structure and transcription of host chromatin in cis, eLife, 2018, 7, e36245.
CRM Bangham, Human T cell leukemia virus type 1: Persistence and pathogenesis, Annual Review of Immunology, 2018, 36, 25–53.
Y Satou, P Miyazato, K Ishihara, H Yaguchi, A Melamed, M Miura, A Fukuda, K Nosaka, T Watanabe, A Rowan, M Nakao, and CRM Bangham, The retrovirus HTLV-1 inserts an ectopic CTCF-binding site into the human genome, Proceedings of the National Academy of Sciences, 2016, 113, 3054–3059.